Design DNA libraries¶
In this section we will generate the primers that are used to do saturation mutagenesis on proteins (ie. NNS primers).
We will also generate each possible point mutant sequence and export it to a Fasta file, which can be useful if you use Twist Bioscience to generate your site saturation library.
- Classes reviewed in this notebook:
Design primers¶
Now we will define the dna sequence, the beginning and end of the mutable part.
from typing import List
from pandas.core.frame import DataFrame
from mutagenesis_visualization import GeneratePrimers
# DNA
dna: str = 'TGTACAGTAATACAAGGGGTGTTATGGAAAAAATTATGCCGGAAGAAGAATACAGCGAATTTAAAGAACTGATTCTGCAGAAGGAACTGCACGTGGTGTATGCACTGAGCCACGTGTGTGGCCAGGATCGTACCCTGCTGGCCAGTATCTTACTGCGCATCTTTCTGCACGAGAAGCTGGAGAGCCTGTTACTGTGCACACTGAACGATCGCGAGATCAGCATGGAAGATGAAGCCACCACCCTGTTCCGCGCAACAACCCTGGCCAGCACCCTGATGGAGCAGTATATGAAAGCCACCGCCACCCAGTTCGTGCATCATGCCCTGAAAGATAGCATTTTAAAAATTATGGAAAGCAAACAGAGCTGCGAACTGAGCCCGAGCAAGCTGGAGAAAAACGAGGACGTGAACACCAACCTGACCCACCTGCTGAACATTCTGAGCGAACTGGTGGAAAAAATCTTTATGGCAAGCGAAATCCTGCCTCCGACCCTGCGTTACATCTACGGCTGCCTGCAGAAGAGCGTGCAGCATAAATGGCCGACCAATACCACCATGCGCACACGTGTGGTGAGCGGTTTTGTGTTCCTGCGTCTGATCTGCCCGGCAATCCTGAACCCGCGCATGTTCAACATCATTAGCGACAGCCCGAGTCCTATCGCAGCACGTACCCTGATCCTGGTGGCAAAAAGCGTGCAAAATCTGGCCAACCTGGTGGAATTTGGCGCCAAAGAGCCGTACATGGAAGGCGTGAATCCGTTTATCAAAAGTAACAAACATCGCATGATCATGTTCCTGGACGAACTGGGCAACGTTCCGGAACTGCCGGATACAACCGAACATAGTCGCACAGACCTGAGTCGTGACCTGGCCGCCCTGCATGAAATCTGCGTGGCCCATAGCGATGAGCTGCGCACACTGAGCAACGAGCGTGGCGCCCAGCAGCACGTGCTGAAGAAACTGCTGGCCATTACCGAACTGCTGCAACAAAAGCAGAACCAGTACACCAAAACCAACGACGTGCGTtatccgtatgatgtgccggattatgcgccatcacttggctagaggcatc'
#^
# Start of protein. Note 'ATG' codon is the first codon.
start: str = 'ATGGAAAAAATTATGCCGGAAGAA'
# The 'tat' codon will be the first codon that is not mutated
end: str = 'tatccgtatgatgtgccggattatgcg'
# Initialize instance of class GeneratePrimers
generate_primers : GeneratePrimers = GeneratePrimers(dna, start, end)
Set all primers to have the same base pair length.
df_primers: DataFrame = generate_primers(codon='NNS', length_primer=15)
Set all primers to have the same melting temperature.
df_primers_tm: DataFrame = generate_primers(codon='NNS', melting_temp=60)
If you just want to export the file to excel. This command must be run after first generating a dataframe.
generate_primers.export_file(output_file="path/to/file.xlsx")
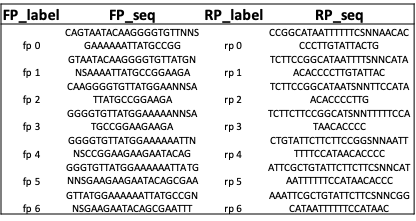
Design site-saturation sequences¶
Define dna sequence and the list of codons that we want to use to generate the mutants.
from mutagenesis_visualization import CreateVariants
# list of codons we want to use
codon_list: List[str] = ["GCC", "GCG", "TGC", "GAC", "GAG", "TTC"]
# DNA sequence we are going to use as the template
dna: str = 'ATGGCCGTGGGGTGTTATGGATGTACAGTAATACAAGGGGTGTTATGGAAAAAATTATGCCGGAAGAAGAATACAGCGAATTTAAAG'
# Initialize instance
create_variants: CreateVariants = CreateVariants()
Get a dataframe with the sequences:
df_variants: DataFrame = create_variants(dna, codon_list)
If you just want to export the file to fasta. This command must be run after first generating a dataframe.
create_variants.export_file(output_file="path/to/sequences.fasta")

If you just want to export the file to excel:
create_variants.export_file(output_file="path/to/sequences.xlsx")